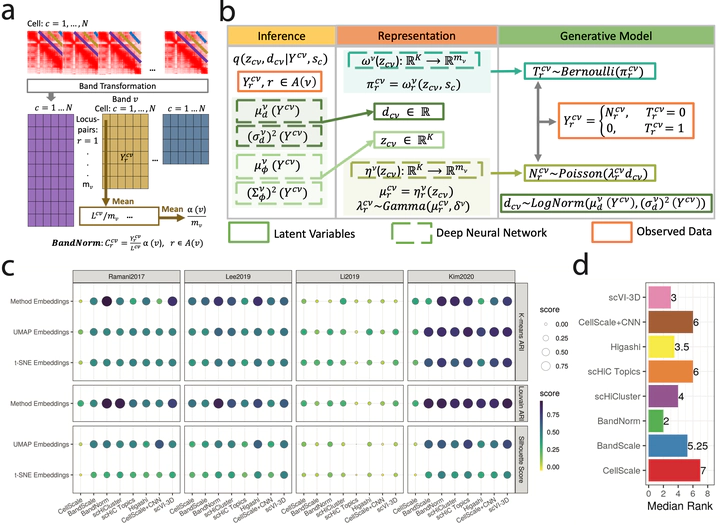
Abstract
Single-cell high-throughput chromatin conformation capture methodologies (scHi-C) enable profiling long-range genomic interactions; however, data from these technologies are prone to technical noise and biases that hinder downstream analysis. We developed a normalization approach, BandNorm, and a deep generative modeling framework, scVI-3D, to account for scHi-C specific biases. In benchmarking experiments, BandNorm yields leading performances in a time and memory efficient manner for cell-type separation, identification of interacting loci, and recovery of cell-type relationships, while scVI-3D exhibits advantages for rare cell types and under high sparsity scenarios. Application of BandNorm coupled with gene-associating domain analysis reveals scRNA-seq validated sub-cell type identification.