Repetitive Regions of Genomes
Project Introduction
Repetitive DNA is nucleotide sequences that appear multiple times across genome which are abundant in a broad range of species, from bacteria to mammals and cover nearly half of the human genome. The role of repetitive DNA in the genome has remained speculative for decades however several recent studies have demonstrated that repetitive DNA can be potentially a vital drive for stabilizing genome contact. For instance, short tandem repeats, whose expansion is the direct reason for more than 25 inherited human disorders such as fragile X syndrome and Huntington, has proven to co-localize with chromatin domain boundaries (Sun et al. 2018. Cell).
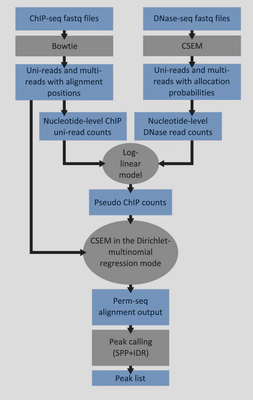
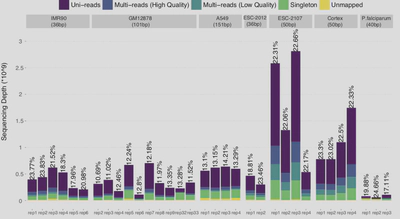
My interest in the repetitive DNA began when I was participating in the ENCODE III project where I developed Permseq (Xin et al. 2015. PLOS Computational Biology), an R package to mapping protein-DNA interactions in highly repetitive regions of the genomes with prior-enhanced read mapping. I have found striking false positives and false negatives if reads originated from the repetitive regions are discarded due to alignment uncertainty.
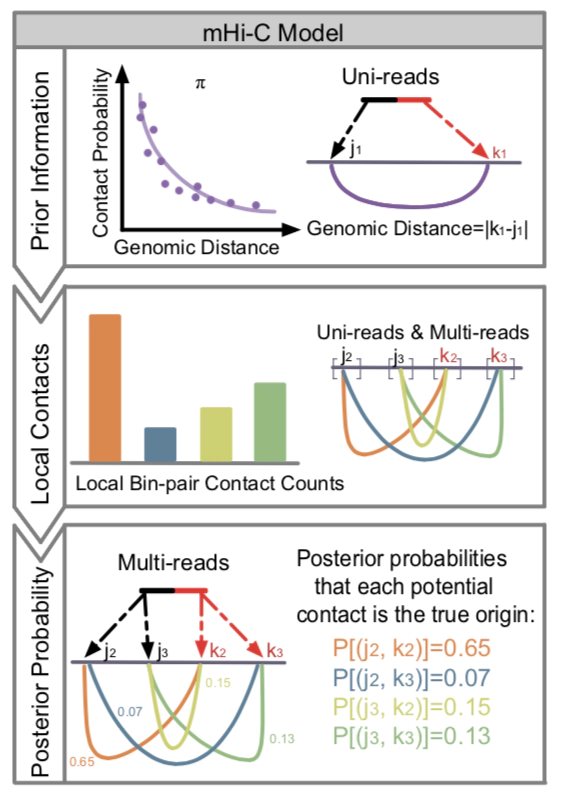
Later on, I developed mHi-C (Zheng et al. 2020. Nature Methods) to address similar multi-mapping reads issue in 3D nucleosome studies yielding significant improvement in sequencing depth and refined inference of genome structure and functions.
Publications
-
Cheng J, Clayton J, Acemel R, Zheng Y, Taylor R, Keleş S, Harley J, Quail E, Gómez-Skarmeta J and Ulgiati D. Regulatory architecture of the RCA gene cluster captures an intragenic TAD boundary and enhancer elements in B cells. Frontiers in Immunology, section B Cell Biology. 2022.
-
The ENCODE Project Consortium, et al. Expanded Encyclopedias of DNA Elements in the Human and Mouse Genomes. Nature. 2020 .
-
The ENCODE Project Consortium, Snyder, M.P., Gingeras, T.R., Moore, J.E., Weng, Z., Gerstein, M.B., Ren, B., Hardison, R.C., Stamatoyannopoulos, J.A., Graveley, B.R., Feingold, E.A. and Pazin, M.J. Perspectives on ENCODE. Nature. 2020.
-
Zheng Y, Ay F, Keleş S. Generative modeling of multi-mapping reads with mHi-C advances analysis of Hi-C studies. eLife. 2019.
-
Zeng X, Li B, Welch R, Rojo C, Zheng Y, Dewey CN, Keleş S. Perm-seq: Mapping Protein-DNA Interactions in Segmental Duplication and Highly Repetitive Regions of Genomes with Prior-enhanced Read Mapping. PLoS Computational Biology. 2015.