CAR-T cell therapy genomic profiling and clinical association study.
Project Introduction
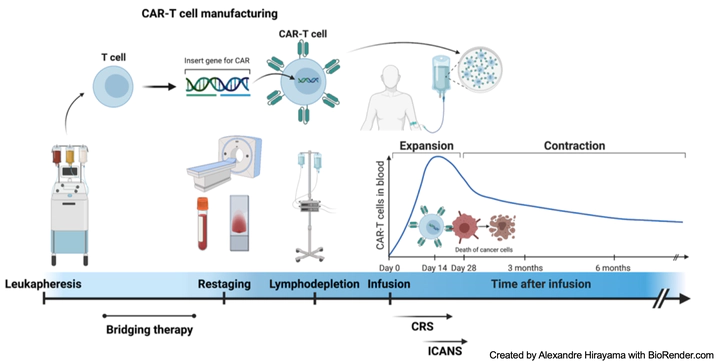
Motivated by computational needs and challenges in infectious diseases and immunotherapy, I have been actively working on the development of statistical methods and tools for analyzing single-cell multi-omics data in addressing immunological and immunotherapeutic problems. My postdoctoral training at the Fred Hutchinson Cancer Center enables me to participate in immunological studies and interact with clinicians directly. This significantly allows me to gain insights into high-throughput assays and high-dimensional cellular analysis, specially designed to address immune cell heterogeneity. Through close collaboration with the Turtle Lab, I have constructed a specific computational framework for processing and analyzing the CAR-T cell therapy CITE-seq data. Additionally, with the Turtle Lab and the Henikoff Lab, we have also profiled the epigenomic landscape (using CUT&RUN, scCUT&Tag data) of the CAR-T cells of both patients and healthy donors to investigate the cell type origin after CAR-T manufacturing stimulation. Collectively, we can reveal the genomic marker association with the CAR-T cell therapy clinical efficacy and toxicity from both transcriptomics and epigenomics perspectives.
I have built a repertoire of collaborative research experience with statisticians, computational biologists, molecular biologists, and immunologists worldwide to address immunological and immunotherapeutic challenges.
Publication
-
Fiorenza S+, Zheng Y+, Sarthy J, Sheih AS, Wu Q, Hirayama AV, Kimble EL, Kirchmeier D, Gottardo R, Henikoff S, Turtle CJ. High-throughput, high sensitivity mapping of human T cell and CAR-T cell epigenomic landscapes underscores the role of H3K27me3 in subset differences and therapeutic outcomes. Manuscript in preparation. (+ Co-first author)
-
Germanos AA, Arora S, Zheng Y, Goddard ET, Coleman IM, Ku AT, Wilkinson S, Amezquita RA, Zager M, Long A, Yang YC, Bielas J, Gottardo R, Ghajar C, Nelson P, Sowalsky A, Setty M, Hsieh A. Defining cellular population dynamics at single cell resolution during prostate cancer progression. BioRxiv. 2022. Under review for eLife.
-
Hirayama AV, Zheng Y, Dowling MR, Sheih A, Phi TD, Kirchmeier DR, Chucka AW, Gauthier J, Maloney DG, Gottardo R, Turtle CJ. Long-Term Follow-up and Single-Cell Multiomics Characteristics of Infusion Products in Patients with Chronic Lymphocytic Leukemia Treated with CD19 CAR-T Cells. Blood. 2021.
-
Vitanza N, Biery M, Myers C, Ferguson E, Zheng Y, Girard E, Przystal J, Park G, Noll A, Pakiam F, Winter C, Morris S, Sarthy J, Cole B, Leary S, Crane C, Lieberman N, Mueller S, Nazarian J, Gottardo R, Brusniak M, Mhyre A, Olson J, Optimal therapeutic targeting by HDAC inhibition in biopsy-derived treatment-naïve diffuse midline glioma models. Neuro-Oncology. 2021.
-
Zheng Y, Ahmad K, Henikoff K. CUTTag Data Processing and Analysis Tutorial. Protocols.io. 2020. (15,266 views, 4,355 exports, and 194 questions)
-
Liao R+, Zheng Y+, Liu X, Zhang Y, Seim G, Tanimura N, Wilson G, Hematti P, Coon J, Fan J, Xu J, Keleş S++ and Bresnick E++. Discovering How Heme Controls Genome Function Through Heme-omics. Cell Reports. 2020. (+co-first authors, ++co-corresponding authors)
-
Soukup AA, Zheng Y, Mehta C, Liu P, Hofmann I, Zhou Y, Zhang J, Choi K, Johnson KD, Keles S, Bresnick EH. Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer. Journal of Clinical Investigation. 2019.
-
Tanimura N, Liao R, Wilson GM, Dent MR, Cao M, Burstyn JN, Hematti P, Liu X, Zhang Y, Zheng Y, Keleş S, Xu J, Coon J, Bresnick E. GATA/Heme Multi-omics Reveals a Trace Metal-dependent Cellular Differentiation Mechanism. Developmental Cell. 2018.